Tumour and normal adjacent tissue pairs in breast cancer.
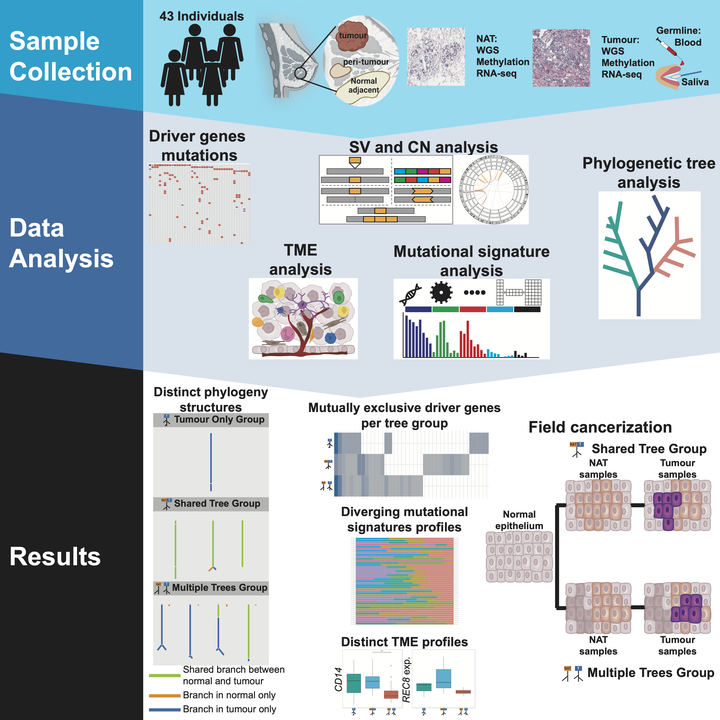
Breast cancer is a complex disease that evolves through dynamic interactions between tumour cells and their surrounding environment. In our study, “Genomes and epigenomes of matched normal and tumour breast tissue reveal diverse evolutionary trajectories and tumour-host interactions” published in AJHF, a collaboration between our group at the University of Manchester and researchers at the National Institutes of Health (NIH) National Cancer Institute (NCI), we focus on studying tumour and normal adjacent tissue (NAT) in breast cancer patients from Hong Kong. Our study reveals complex genetic and epigenetic interactions in both tumour tissue and NAT.
The initial step of our analysis focused on identifying genomic and epigenetic characteristics in our samples, including driver genes, tumour microenvironment (TME) compositions, and mutational signatures within each sample. This allowed us to characterise the differences between the NAT and tumour sample groups. We then processed the NAT-tumour sample pairs in tandem using Multisample DP-Clust, a tool developed by our lab, in order to obtain a holistic view of the different evolutionary trajectories of breast cancer.
Our analysis identified three distinct evolutionary patterns:
- Shared-Tree Group: This trajectory reflects shared evolution between normal tissue and tumours, indicating that genetic mutations occurred early and affected both tissues.
- Multiple-Tree Group: This trajectory shows independent evolution between normal tissue and tumours, with separate genetic trajectories.
- Tumour-only Tree Group: This trajectory reveals mutational evolution occurring exclusively in the tumour samples.
Subsequently, we explored the unique characteristics within each trajectory group to understand the evolutionary trajectory in each group. Interestingly, these differences were linked to how the tumour interacts with its environment. The study reveals significant variation in the tumour and NAT between the three groups. For instance, NAT in the multiple-tree group tends to be less inflammatory, exhibiting higher levels of regulatory T cells and fewer CD14+ immune cells. Additionally, we observed differences in driver gene mutations and mutational signature composition across the three groups. Tumours and NAT samples in the multiple-tree group harboured more aggressive driver gene mutations. This less inflammatory immune landscape could influence tumour growth and response to treatments.
Our findings support the concept of “field cancerisation”, where NAT harbours genetic changes independent of the tumour, and reinforces the idea that the tumour microenvironment plays a crucial role in cancer development, not just the tumour itself.
These findings underscore the complexity of breast cancer evolution, shaped by both intrinsic genetic factors and external interactions with the tumour microenvironment (TME). By understanding these diverse evolutionary dynamics, we can develop more effective therapies that target both the tumour and its surrounding environment. This brings us closer to personalised treatment approaches for breast cancer, where both the genetic landscape and tumour microenvironment are considered.
Our work has been published in Volume 111, Issue 12 in AJHG and is also available online.